Over the last few decades, the scientific community has turned its gaze towards a promising field in the fight against cancer – immunology. The idea of harnessing the power of the immune system and functional cytokines to develop cancer treatments has been gaining traction.1 However, the cancer immunotherapy development pipeline is a long and winding path that can be full of roadblocks. This complexity stems from the uniqueness of each immune cell and of each patient, which necessitates characterizing individual immune cells to maximize success in the clinic.2
Recent studies have shed light on the importance of polyfunctionality, a characteristic that enables a single immune cell to secrete multiple cytokines. This attribute, while rare, is a determining factor of cellular strength and functionality. In cancer immunotherapy specifically, multiple cytokines secreted by polyfunctional cells work in tandem to directly induce tumor death or modulate immune responses against cancer. 3
To identify and characterize these potent polyfunctional cells, a technology is needed that can measure highly multiplexed cytokines secreted from live single cells. By assaying cellular functionality through cytokine secretion, we can gain valuable insight into therapeutic efficacy.3
Key Steps in Cancer Immunotherapy Development
The journey of developing cancer immunotherapies typically involves 3 critical stages:
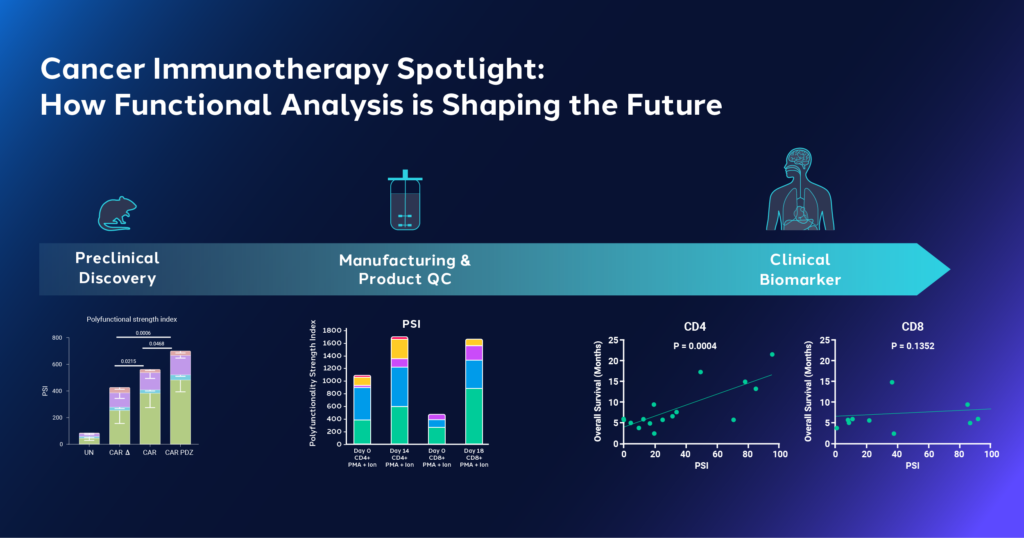
- Preclinical stage: By using in vitro and in vivo animal models, researchers can explore and test modifications, techniques, and designs. This stage is crucial to identifying promising targets and treatment approaches, as well as characterizing their function. 3
- Manufacturing optimization: Achieving consistency, scalability and functionality are key during this stage, which often involves exploring techniques and compounds that can preserve functional phenotypes. 3
- Clinical studies: This stage is necessary to evaluate the performance of the ACT in humans, particularly because donor heterogeneity can result in product and response heterogeneity, necessitating a deeper understanding of ACT performance through clinical biomarkers. 3
Spotlight on Bruker Cellular Analysis’ Technology: Real-World Case Studies Across Preclinical, Manufacturing, and Clinical Stages of Cancer Immunotherapies
Preclinical Characterization
A study in Nature Biotechnology leveraged Bruker Cellular Analysis technology to characterize the functional efficacy of CAR therapies for solid tumors. Rather than finding new cancer cell targets, researchers focused on improving CAR binding to antigens. This was achieved by strengthening the synapse between CAR and antigens and adding an anchor domain from CRTAM, a protein involved in synapse formation, to CARs. This created a modified CAR design known as CAR.PDZ which was hypothesized to demonstrate stronger binding, quicker activation, and increased potential for cytotoxic activity.4
The study used the CAR.PDZ design to create CAR Natural Killer (NK) cells. Utilizing IsoCode® Single-Cell Secretome platform, researchers found that CAR.PDZ NK cells had higher polyfunctionality and polyfunctional strength index® (PSI®) compared to other control CAR-NK and NK cells. Single-cell data analysis revealed distinct groups formed by CAR.PDZ NK cells, indicating the modification’s impact on NK cell function. Further experiments showed improved survival and increased cytotoxicity in lung cancer and osteosarcoma mouse models treated with CAR.PDZ NK cells. 4 These findings suggest that this modification confers enhanced efficacy for CARs and highlights the importance of single-cell functional phenotyping in understanding cell modifications. 4
In another study published in Communications Bio, researchers established a method using Opto-Electro Positioning (OEP®) technology on the Beacon® platform to improve the identification and purification of properly edited clones, a process that is still difficult despite CRISPR advancements. 5 The technique allows for single cell-manipulation on an OptoSelect® chip, enabling the assessment of single-clone editing efficiencies within 10 days. 5 In particular, OEP on the Beacon provided researchers with the ability to interrogate T cell functionality by assessing proliferation and surface labeling indicating successful gene editing. Functional cells were then exported for downstream applications. The ability to determine the phenotype individual cells after CRISPR modification can be used to evaluate gene editing strategies while the export capabilities allow for further functional analysis of successfully modified cells.
Manufacturing Optimization
In research published in Frontiers, a team at Lonza developed a scalable manufacturing platform for producing allogeneic T cells.6 Due to the high T cell dose per patient and growing patient numbers, there is growing clinical demand for these allogeneic cells, necessitating a scalable production platform that can maintain cell quality.
Using Bruker Cellular Analysis’ IsoCode ® Single-Cell Secretome platform, the team assessed the function of peripheral blood mononuclear cells (PBMC)-derived T cells before and after expansion using 3D suspension in stirred tank bioreactors. After expansion, T cells showed increased polyfunctionality after stimulation, and an increased frequency of polyfunctional cells secreting 5+ cytokines. Both subsets of T cells, CD4+ and CD8+, exhibited an increase in effector polyfunctional strength index® (PSI®) after expansion, with a notable increase in stimulatory PSI. 6 This study highlights that stirred tank bioreactor culture enhances polyfunctionality and cytokine secretion potency of T cells and may be used as a scalable platform for T cell manufacturing to meet clinical demand.
Clinical
In a study published in Blood Advances, researchers from MD Anderson used Bruker Cellular Analysis’ platform to explore the role of T cell function in predicting responses to immune-based treatments in Acute Myeloid Leukemia (AML) patients.7 They performed a multiplexed immune assay to assess the functional states of CD4+ and CD8+ cells at a single-cell level in pretherapy bone marrows from 16 AML patients treated with azacitidine/nivolumab. 7
The study found that effector CD4+ cells, but not CD8+ cells, exhibited distinct polyfunctional groups linked to better responses and outcomes. This polyfunctionality was primarily driven by interferon-gamma (IFN-g) and tumor necrosis factor-alpha (TNF-a). Identifying cellular subsets and specific cytokine profiles in responder cohorts provides clinicians with improved understanding of therapeutic performance. The team concluded that these single-cell polyfunctional assays could predict AML response and potentially serve as a biomarker in immunotherapy, which could be useful for treating AML, which remains a challenging disease to manage.7
The Growing Impact of Bruker Cellular Analysis’ Functional Analysis Instruments on Cancer Immunology
The world of cancer immunology is being reshaped by the power of functional analysis and the innovative technology of Bruker Cellular Analysis’ single-cell analysis platforms. The IsoSparkTM proteomic barcoding instrument and the Beacon® Optofluidic platform are creating a paradigm shift in how we understand and approach cancer treatment.
Functional analysis has proven to be an invaluable tool, offering a detailed picture of cellular responses to immune-based therapies. It provides a unique lens through which researchers can view and predict patient responses, tailoring therapies to meet individual needs and increasing their effectiveness. Bruker Cellular Analysis’ single-cell analysis platform takes this a step further. It allows researchers to delve deeper into the cellular universe, identifying rare subsets of highly polyfunctional cells associated with improved outcomes in patients.
References
- https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3763400/
- https://brukercellularanalysis.com/research-areas/cancer-immunology/
- https://brukercellularanalysis.com/resource/characterizing-adoptive-cell-therapies-using-bruker-cellular-analysis-proteomic-barcode-technology
- https://www.nature.com/articles/s41587-022-01650-2
- https://www.nature.com/articles/s42003-018-0034-6
- https://www.frontiersin.org/articles/10.3389/fmedt.2022.850565/full
- https://pubmed.ncbi.nlm.nih.gov/34555853/