While genomic sequencing has been instrumental in revealing genetic mutations that promote the development of cancerous cells, additional research techniques are necessary to fully understand how gene mutations affect cell function and signaling networks, in turn helping researchers uncover the significant mechanisms driving carcinogenesis. The complexity of intracellular signaling in cancer cells makes it difficult to ascertain what signaling pathways are dysregulated or highly activated; furthermore, uncovering these pathways requires the ability to analyze numerous phosphoproteins at single-cell resolution, at the same time. Bruker Cellular Analysis’ Single-Cell Signaling platform uncovers coordinated signals with its unique ability to measure 15+ phosphoproteins simultaneously per single cell to help researchers elucidate mechanisms of tumorigenesis.
Understanding the Link between Gene Mutations and Cancer
In a paper recently published in Nature Genetics, researchers analyzed samples from patients with chronic lymphoproliferative disorder of natural killer cells (CLPD-NK), a type of leukemia with poorly understood genetic causes that is characterized by a persistent increase of mature NK cells in the blood. In the study, researchers discovered that the gene CCL22 was mutated in a subset of patients and had not been previously identified in CLPD-NK. CCL22, encoded by this gene, is a chemotactic protein that recruits immune cells expressing the chemokine receptor, CCR4. When wild type CCL22 binds with CCR4, it triggers downstream signaling pathways and causes the CCR4 receptor to be internalized, halting excessive signaling and thus, harmful carcinogenesis.
To better understand how the mutation affects cell function and then leads to carcinogenesis, the authors incubated Ba/F3 cells (murine B cells) expressing wild-type CCR4 (Ba/F3-CCR4) with wild-type or mutant CCL22. They found that the mutant CCL22 increased the chemotaxis, or migration, of cells relative to the control. Furthermore, they found that mutant CCL22 impaired the internalization of the CCR4 receptor, amplifying the effects of CCL22 and preventing the cessation of excessive signaling.
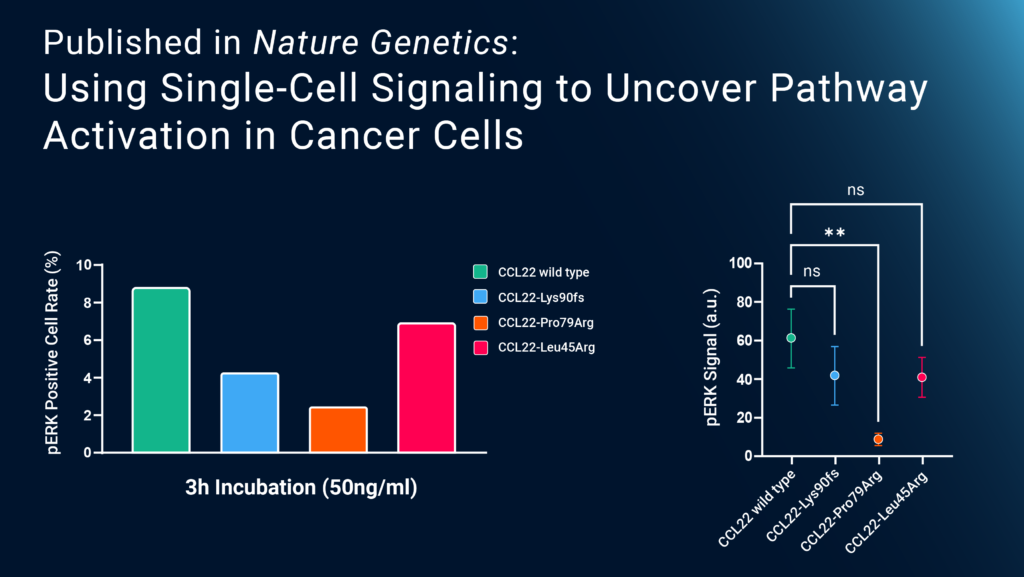
To see how this affected intracellular signaling, the study used Bruker Cellular Analysis’ Single-Cell Signaling platform for highly multiplexed phosphoproteomic analysis. They loaded murine Ba/F3-CCR4 cells pretreated with wild-type or mutant CCL22 onto Bruker Cellular Analysis’ human tumor signaling chips and found that the mutant CCL22 decreased phosphorylation of ERK, a kinase associated with the mitogen-activated protein kinase (MAPK) pathway, but not Akt, a common oncogene. Additional experiments demonstrated that mutant CCL22 not only can drive NK cell proliferation in vivo, but also contributes to dysregulation of the microenvironment. Together, these results show, for the first time, how a mutation of the CCL22 gene leads to abnormal NK cell proliferation and, ultimately, the development of CLPD-NK. Bruker Cellular Analysis’ Single-Cell Signaling data helped highlight the dysregulated intracellular pathway that could be a promising target for future CLPD-NK treatments.
Using Single-Cell Phosphoproteomics to Uncover Cellular Function
Multiplexed analysis of the phosphoproteome at the single-cell level helps researchers explore multiple intracellular signaling pathways within each cell, giving researchers a better understanding of how individual cells are functioning and communicating with each other. Bruker Cellular Analysis’ single-cell analysis platform provides deep insights into cell function and signaling network, enabling researchers to identify novel druggable targets for improved targeted therapies.