Understanding Tumor Immunology Across Space and Time
A persistent challenge in tumor immunology is that our most powerful tools tend to capture biology in fragments. Live single-cell assays can tell us what immune cells do, while spatial biology reveals where cells are within intact tissue. But rarely are these two views connected. As a result, researchers are often left inferring function from static phenotypes or interpreting spatial maps without knowing which cells are truly tumor reactive. Fully understanding anti-tumor immunity requires linking live T cell function with spatial biological context. Only by connecting these dimensions can we explain why some T cells successfully engage tumors while others remain ineffective despite appearing phenotypically similar.
This work highlights a close collaboration between Bruker Cellular Analysis, Bruker Spatial Biology, and Dr. Joseph Zenga and Dr. Tyce Kearl at the Medical College of Wisconsin to address this challenge in head and neck cancer. In this study, patient-derived tumor-reactive T cells were first functionally characterized using the Beacon® Discovery platform, enabling direct measurement of live single-cell behavior and identification of antigen-specific responses (Figure 1 top). Spatial biology analysis was then performed from same-patient tumor biopsies using the CosMx® platform to map these functional phenotypes back into intact tumor tissue (Figure 1 bottom). By combining live functional profiling with high-plex spatial context, this integrated approach delivers deep biological insight into how tumor-reactive T cells function, localize, and interact within the tumor microenvironment.
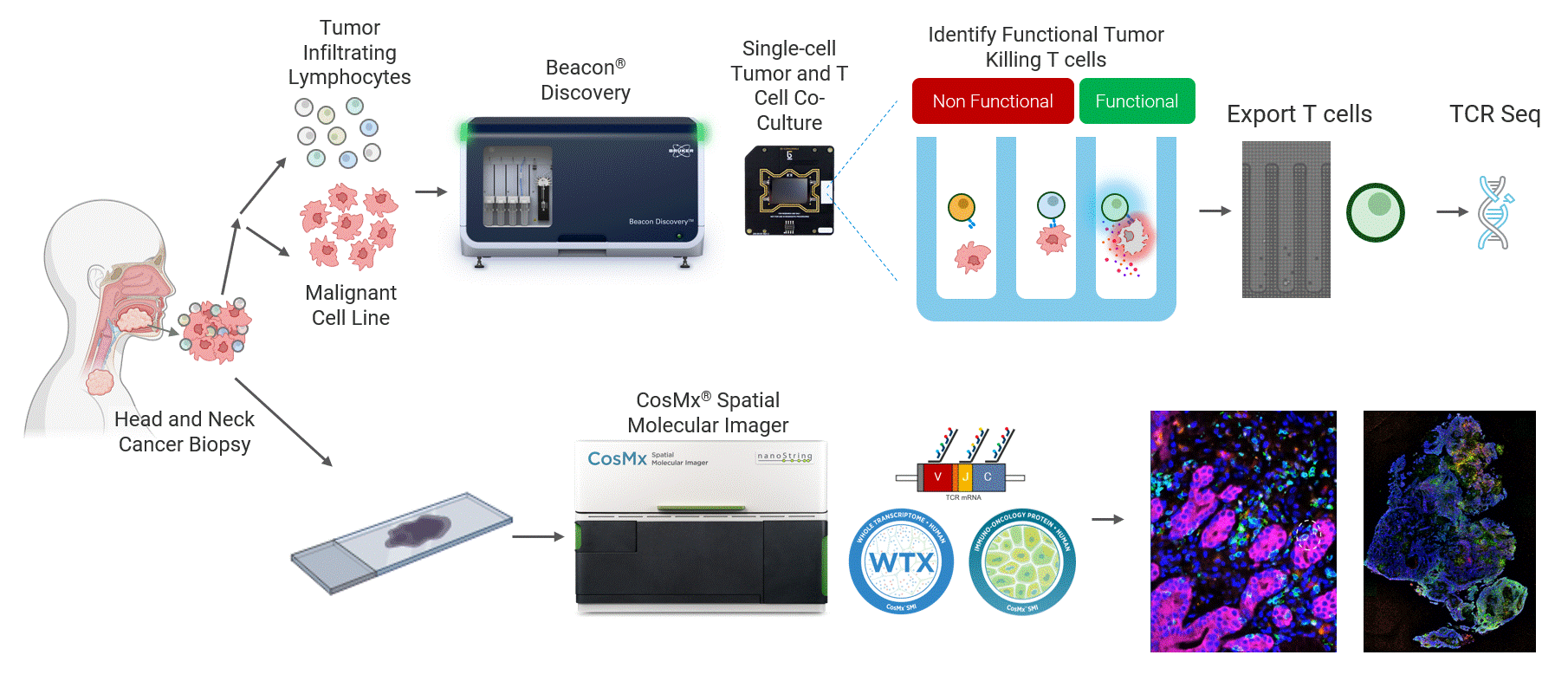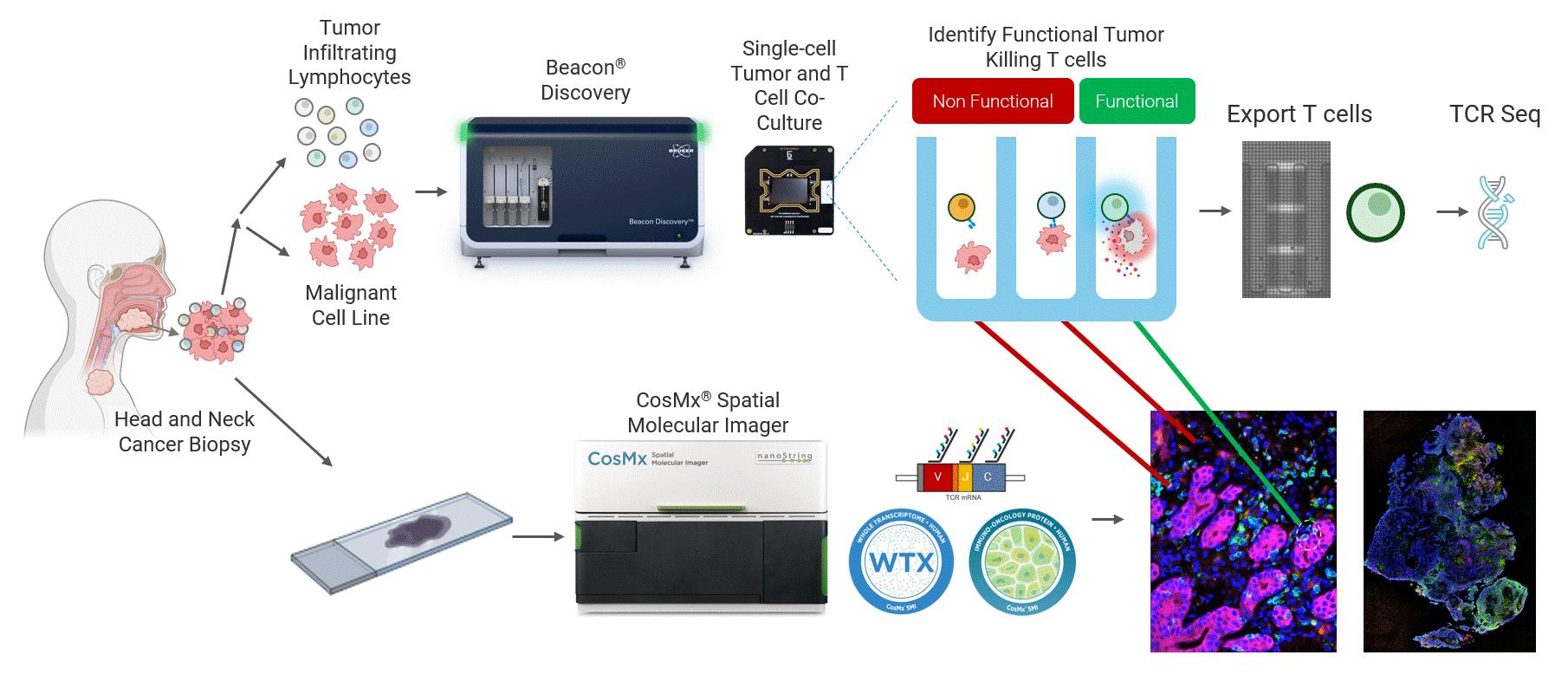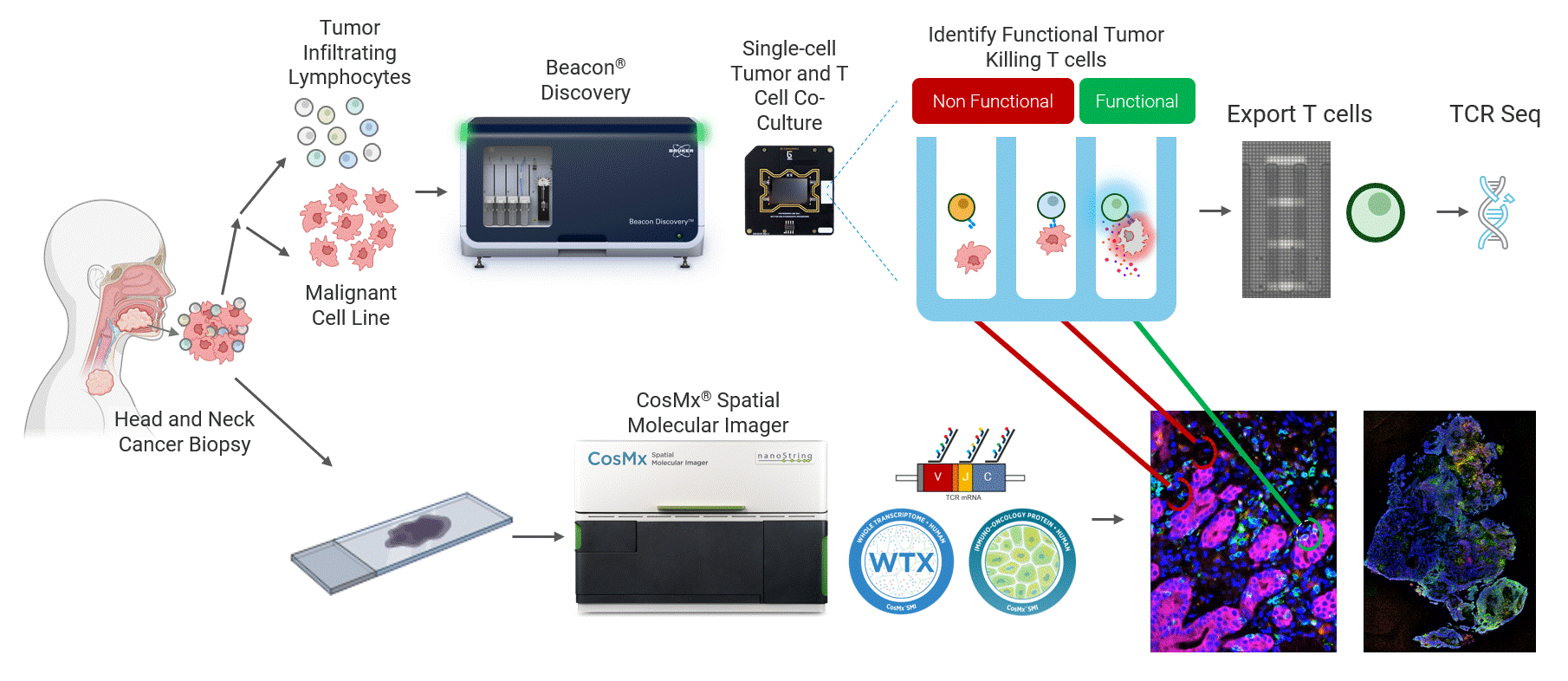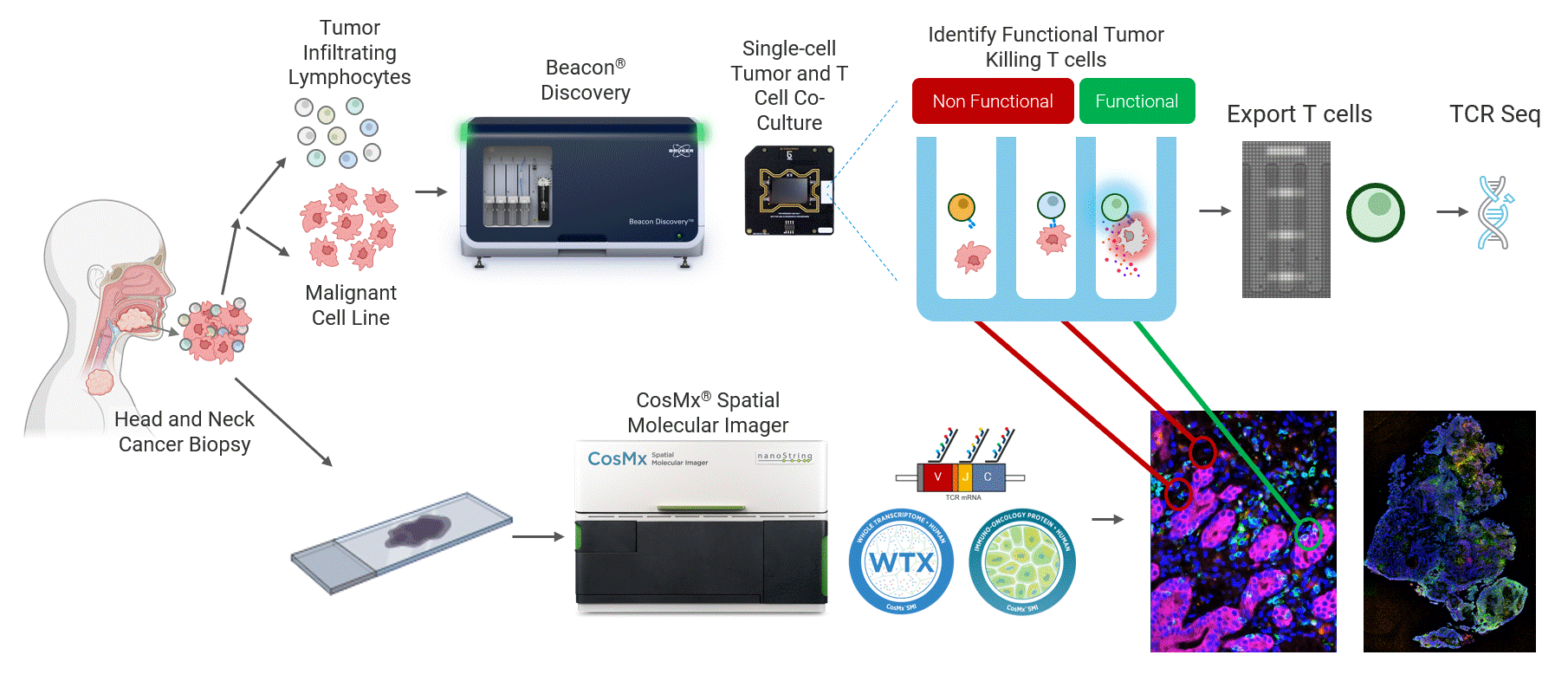
Figure 1 Head and neck cancer tumors are dissociated and co-cultured on the Beacon platform to functionally identify tumor-reactive T cells, which are then exported for downstream sequencing (top). In parallel, head and neck cancer biopsies from the same patient are analyzed on the CosMx platform (bottom) using a targeted TCR panel, whole transcriptome (WTX), and an immuno-oncology protein panel to spatially resolve tumor-reactive T cells within the tumor microenvironment.
Live Single-Cell Analysis to Identify Tumor-Reactive T Cells
The Beacon platform excels at live functional analysis of individual T cells. By establishing single-cell co-cultures and functional assays, the Beacon platform provides a real-time view of antigen-specific T cell behavior, including cytotoxicity, cytokine secretion, and dynamic cell–cell interactions. This function-first approach allows researchers to rapidly identify tumor-reactive T cells based on what they do rather than relying on surrogate markers [1, 2]. (Figure 2)
Critically, these same live cells can be exported and sequenced, enabling recovery of the TCRs associated with true functional activity. This creates a direct link between functional potency and TCR identity, forming a foundation for downstream discovery, validation, and clinical translation applications.
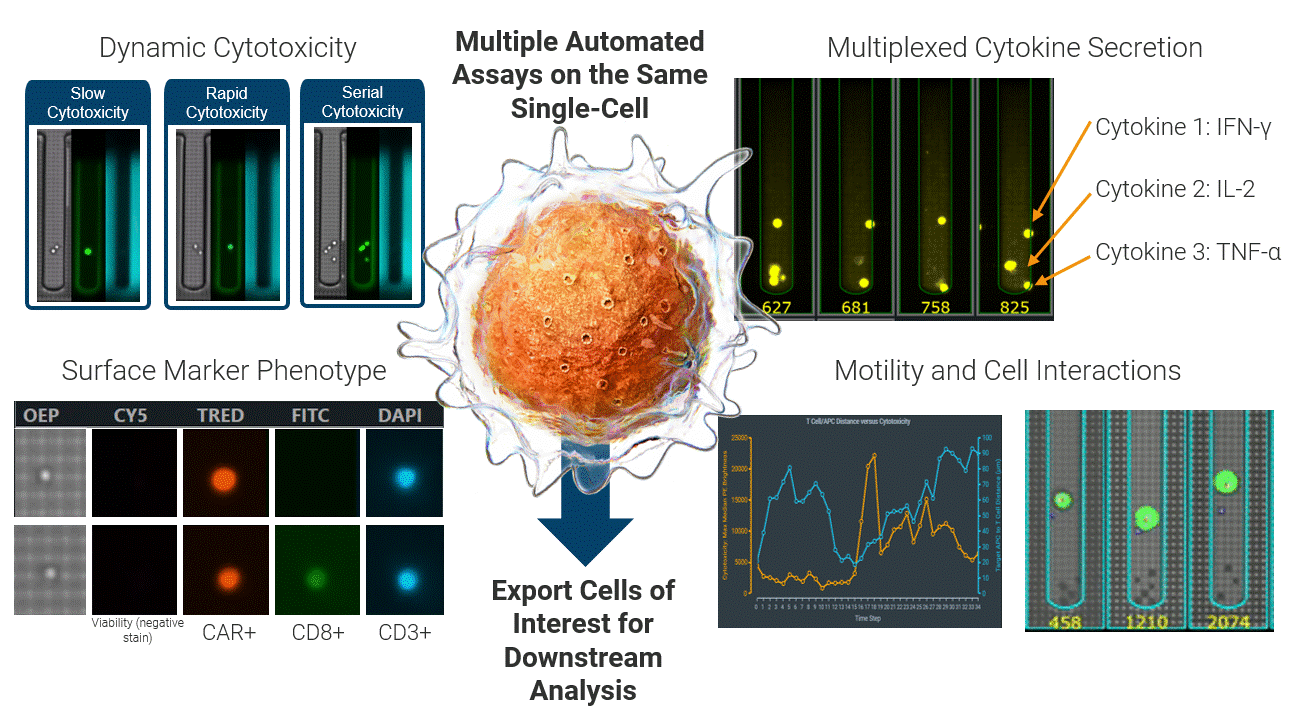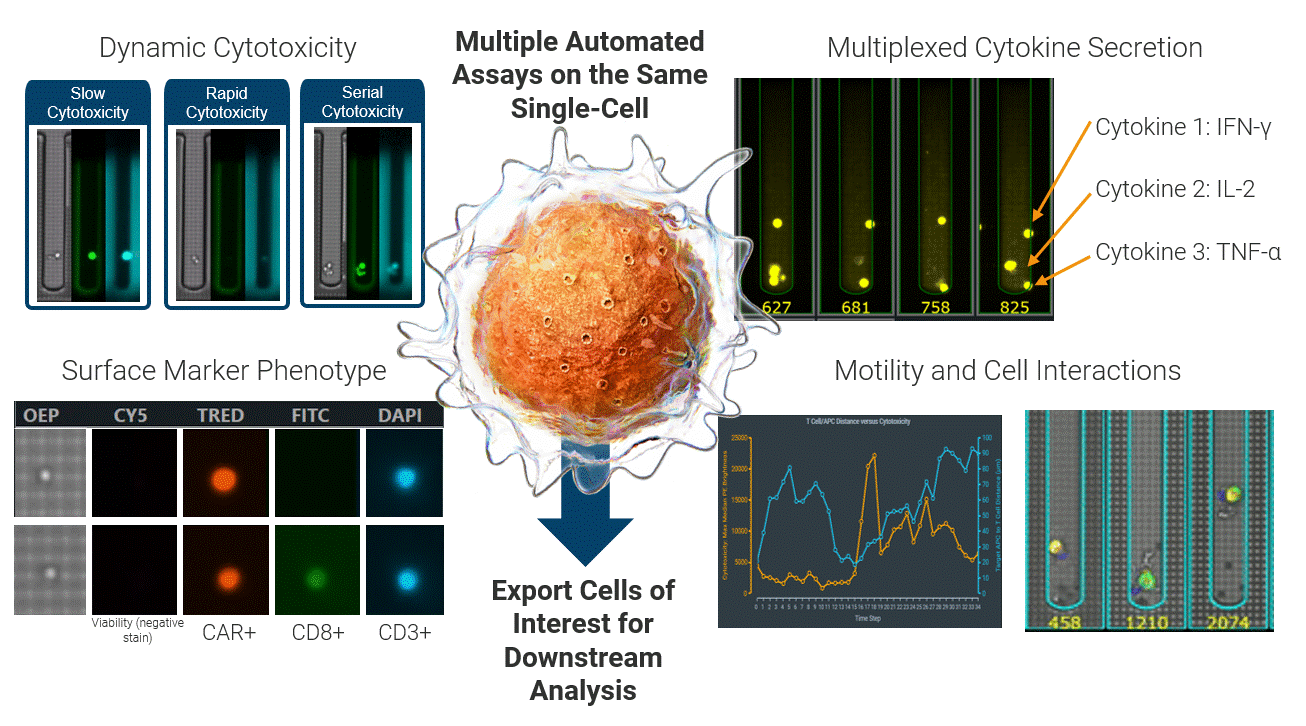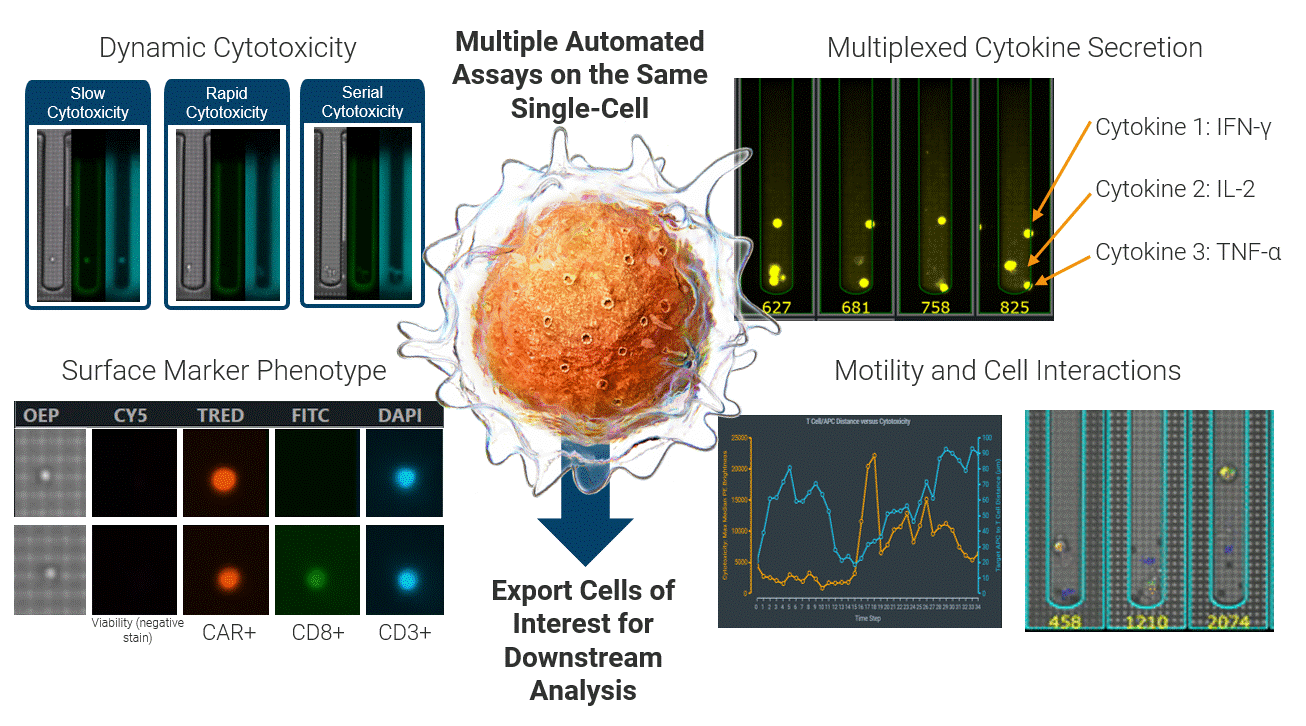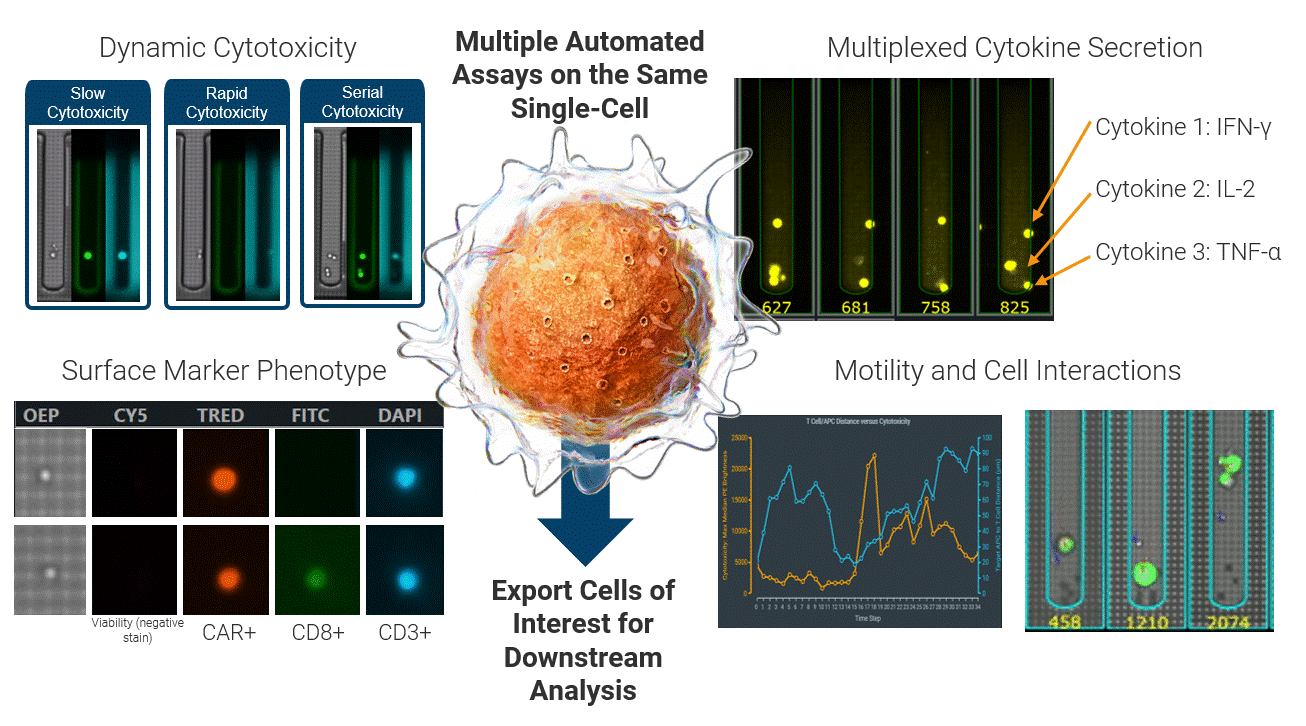
Figure 2 Integrated single-cell functional profiling on the Beacon platform enables simultaneous measurement of cytotoxicity, cytokine secretion, surface phenotype, and dynamic cell behavior from the same individual cell. Functional cells can then be exported for TCR sequencing, directly linking live function to TCR identity.
Mapping Functional TCRs Back into the Tumor Environment
While live functional profiling reveals which T cells matter most, spatial biology provides the missing tissue environment context[3]. Using an early-access targeted TCR panel on the CosMx platform, functional TCRs discovered on the Beacon platform can be identified within fixed tumor tissue. Combined with WTX and Human IO protein panels, this approach delivers a comprehensive view of the tumor microenvironment surrounding those functional clonotypes (Figure 3).
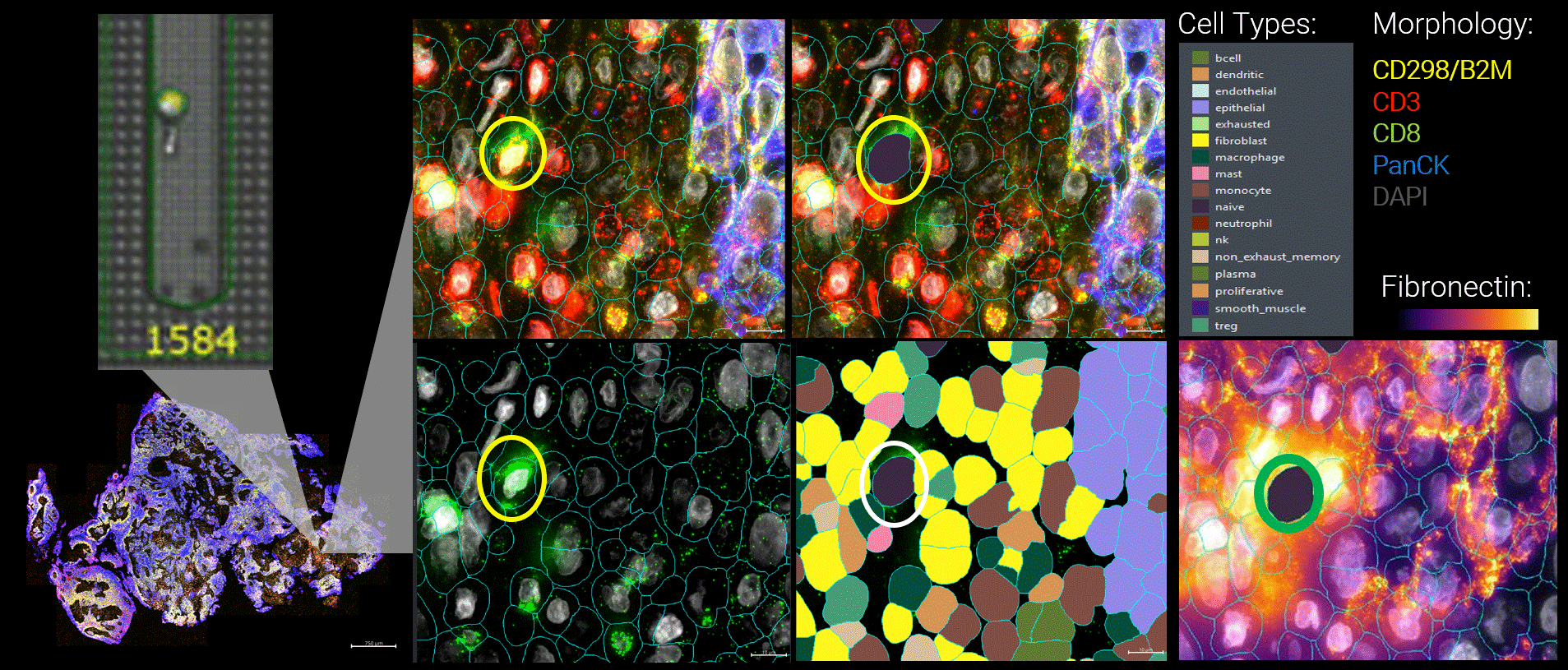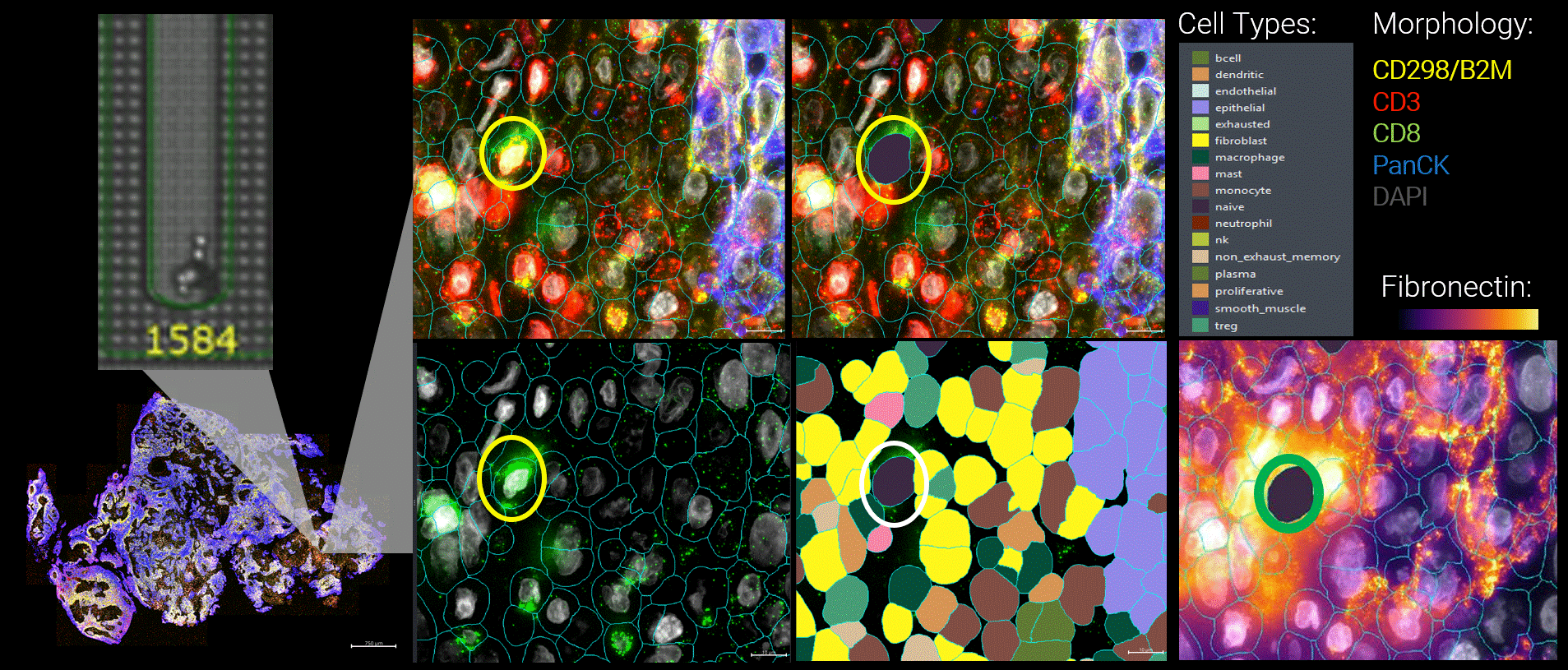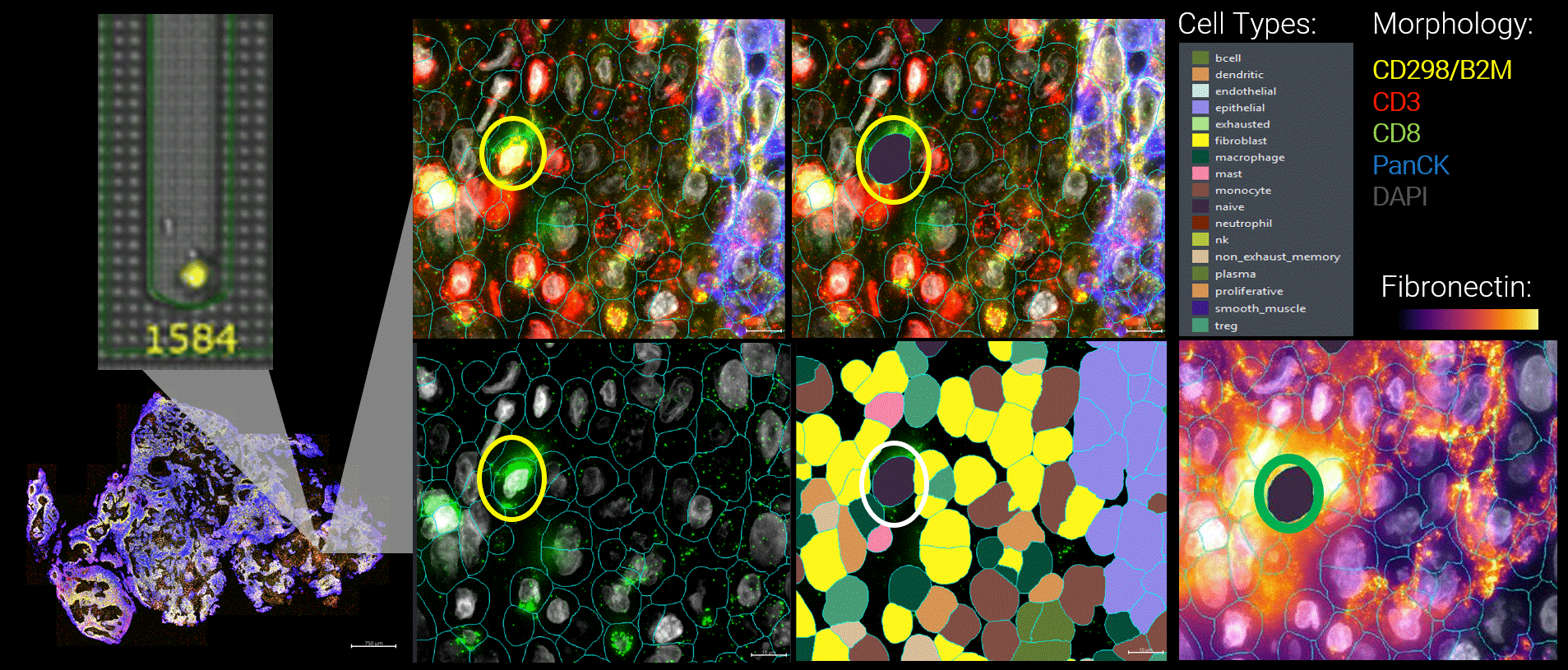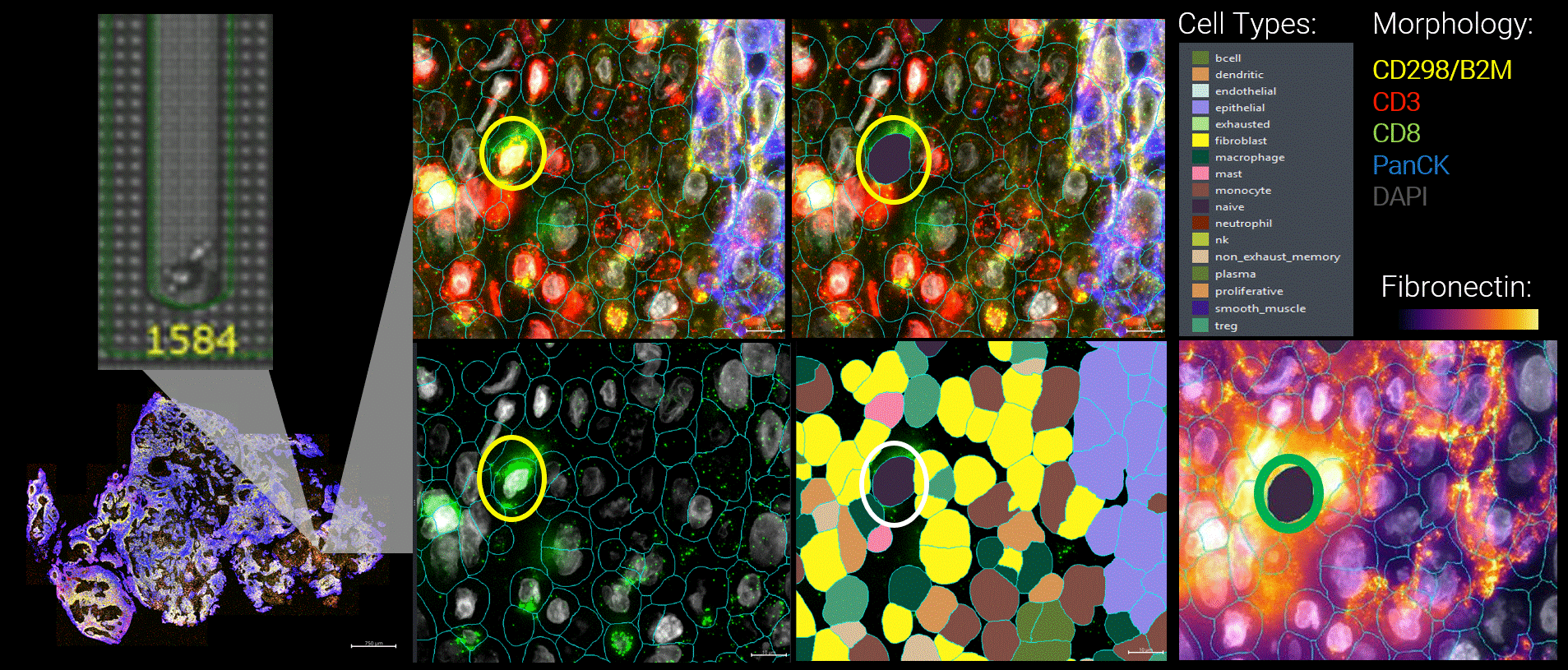
Figure 3 By integrating live functional and spatial data, a more nuanced picture of the tumor microenvironment emerges. Here, functional tumor-infiltrating lymphocytes identified by the Beacon platform were observed infiltrating malignant tumors. Yet despite their functional capacity, these cells were embedded within a suppressive local environment. Regulatory T cells were enriched nearby, and fibronectin encapsulation formed a physical and biochemical barrier around the functional T cell, effectively isolating it within the tumor.
This enables direct spatial localization of tumor-reactive T cells while simultaneously profiling neighboring immune populations, stromal components, and tumor cells. In parallel, live functional data can also be linked indirectly to spatial biology by leveraging single-cell RNA sequencing to define functional phenotypes associated with Beacon platform-identified TCRs and mapping those signatures within tissue. Together, these approaches connect live function to spatial organization through both direct clonotype tracking and phenotype-based inference (Figure 4).
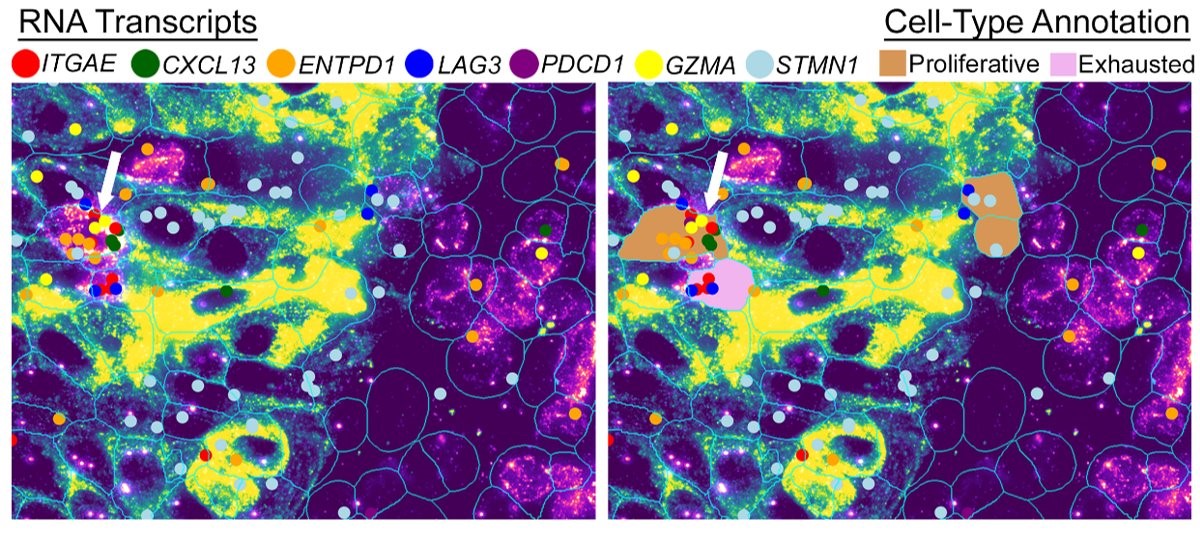
Figure 4 WTX analysis on the CosMx platform reveals the spatial presence of proliferative tumor-reactive T cells alongside exhausted T cell populations within the tumor microenvironment, enabling identification of distinct gene expressions associated with functional state.
These findings help explain why certain therapeutic strategies alone may be insufficient. TIL or TCR therapies may successfully introduce functional cells, and checkpoint inhibitors may relieve some inhibitory signaling, but neither directly addresses physical barriers or suppressive stromal remodeling that limit durable responses.
Furthermore, preliminary results across two patients reveal markedly different immune phenotypes. In one patient, functional tumor-reactive T cells can infiltrate the tumor, yet their activity appears constrained by mechanical barriers such as fibronectin within the surrounding stroma. In the second patient, there is a clear lack of functional T cell infiltration into the tumor microenvironment altogether. Despite sharing the same diagnosis, these patients exhibit fundamentally different underlying biology, highlighting how next-level functional and spatial data can inform the need for distinct therapeutic strategies.
Informing the Next Generation of Therapeutic Strategies by Synergizing Live, Functional Biology with Spatial Biology
This integrated view of live function and spatial context naturally guides next steps. Rather than focusing solely on enhancing T cell potency, data like this point toward complementary engineering strategies aimed at weakening stromal barriers, inhibiting tumor “healing” or fibrosis responses, and improving T cell infiltration and migration. Importantly, this raises new questions that can now be quantitatively addressed: how frequently do these suppressive architectures form around functional cells, and how do they vary across patients and tumor types?
Together, the Beacon platform and CosMx platform form a uniquely synergistic pairing for tumor immunology by connecting live, temporal function with space. The Beacon platform delivers live single-cell functional data, captures cell–cell interactions and dynamic behaviors over time, and enables export and sequencing of the most relevant cells. The CosMx platform enables whole-transcriptome, single-cell spatial biology with unmatched accuracy, sensitivity, and genomic breadth, delivering same-cell multiomics in intact tissues at single-cell and subcellular resolution.
By linking these capabilities, researchers gain a full, multidimensional understanding of anti-tumor immunity. Not just which cells are present, but which cells function, where they reside, what surrounds them, and how that environment shapes outcomes. This integrated approach moves the field closer to differentiating patient responses based on both functional and spatial biology to design therapies that deliver the right cell, in the right place, at the right time.
References
-
- [1] Wagner, T.R., et al., Conserved programs and specificities of T cells targeting hematological malignancies. bioRxiv, 2025: p. 2025.08.07.668919.
- [2] Zhang, W., et al., Identification of antigen-specific functional CD8<sup>+</sup> T cells using an optofluidic system independent of epitope information. iScience, 2025. 28(10).
- [3] Janesick, A., et al., High resolution mapping of the tumor microenvironment using integrated single-cell, spatial and in situ analysis. Nature Communications, 2023. 14(1): p. 8353.


