“If you want to advance human health and you’re not working towards AGI [Artificial General Intelligence]/ASI [Artificial Super Intelligence] right now the 2 most important things you can be building are enabling hardware and/or scientific data sets/reliable biological data that no one else can or will create. All the ‘AGI will solve cancer’ claims are bull$&*# in the near term until we nail these, and we will…We are all about to be given god-like powers of analysis, [data] foundations will matter.”
– Ben Woodington, x.com, 2/5/2025 https://x.com/WoodingtonBen/status/1887060297254310078
Ben’s right. AI will help us “solve” cancer, and yeah, we’ll figure it out. That’s what we do. But let’s cut through the hype. If curing disease is the goal — and I’m betting that’s the game we’re all playing — then the foundation is simple: sequence data LINKED to functional outcomes.
Training AI Right: We Need Functional Data, Not Just Sequences
Sequencing data is the entry point. It digitizes biology, feeding AI models. But to make those AI models work, we need functional data: “Did this sequence cure the disease?” No shortcuts. No guesswork. A 1-to-1 relationship between sequence and function. That’s what we need. Right now, that data is rare. And I don’t mean grabbing sequencing from one study and functional data from another. I’m talking about this sequence did this function. Period. We’ve seen progress. The software world stepped up and gave us the “AlphaFold Moment” by folding proteins and enabling virtual binding analysis. Huge deal — no question. Nobel-level work. But why don’t sick people have the therapies they need? It’s not because the models aren’t smart enough. It’s because the training data isn’t good enough. Garbage in, garbage out. Full stop.
Biology Has the Answers—We Just Need Access
Here’s the deal: biology already knows how to cure most diseases. The immune system is the most advanced computational engine we’ve got — millions of years of R&D already baked in. You toss an antigen into a biological “computer” (mouse, llama, human, whatever), and it crunches the problem for you. Ignore biology at your own risk. That “low-tech” biological computer is keeping you alive right now. It can do things today that today’s AI models can’t touch — like curing diseases. To find an antibody or cell-based therapeutic we pull cells from an immunized animal, we test their FUNCTION: blocking, killing, whatever the job is. Find a winner? Recover the sequence. Then you put that sequence to work in sick animals (or people) and see if it cures the disease. This is how therapeutics are developed and it’s all about FUNCTION linked to sequence. And here’s the kicker: the training data we need already exists. Eight billion people on this planet. Every one of us is sitting on a goldmine of biological information. People get sick. They get better. The data is right there — we just can’t access it yet.
Unlocking the Data AI Models Need to Succeed
So how do we get sequence-to-function data? Two ways. First, a top-down approach: track immune responses in real people—during sickness, recovery, and treatment. Monitor immune responses and outcomes. Link therapies to FUNCTION. It’s not unethical; it’s smart. Second, a bottom-up approach: start with cells. They’re the building blocks of life. And we’ve got tools to do this. At Bruker Cellular Analysis, we built one of those tools: the Beacon platform. It links sequence to function at the single-cell level. Fully automated. Scalable. Repeatable. And it tests a cell’s function in real-time. This is exactly the kind of data AI systems need.
Is single-cell multiomics the magic bullet? No. But it’s a hell of a start.
Where Are We Today?
The world is quickly trying to address the gap in sequence to function by sharing large datasets. Vevo Therapeutics recently released a massive dataset comprising of “100 million [single] cells and 60,000 experiments, mapping 1,200 drug treatments across 50 different tumor models”1. While this provides comprehensive insights into transcriptional changes, the dataset does not include direct functional data such as protein activity assays or phenotypic measurements. Tahoe-100M offers extensive transcriptomic information without incorporating additional functional data. A lot of data, but still not the kind we desperately need.
Staring Down a Cellular Glacier, Not Just an Iceberg
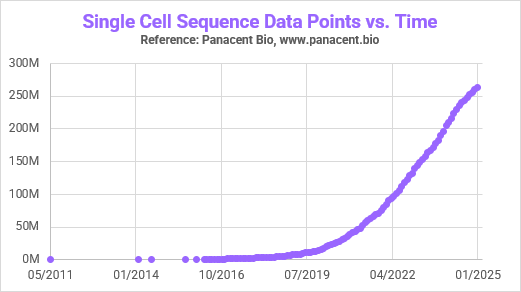
I checked in with my friends Robert Sebra, CEO and Daniel Charytonowicz, CSO of Panacent Bio who have built a comprehensive cellular and molecular foundation model and estimate that, although there are approximately 300 million single cells sequenced in the public domain, only a single digit percentage have functional/perturbational data linked to the sequencing data. So, if we assume 5%, that means <12 million cells have sequence linked to function. For perspective: there are 8 billion humans with 37 trillion cells each. We’re not just at the tip of the iceberg — we’re staring at a glacier the size of Antarctica.
The way forward? Link top-down data with single-cell insights. Build hypotheses with AI. Validate with single cells or multi-cell systems. Build trust. No one’s putting an AI-generated therapy into humans without proving the mechanism works.
At Bruker Cellular Analysis, we’re pushing hard to make this happen. This is the frontier. And it’s an incredible time to be alive. Let’s move faster.


